class: center, middle, inverse, title-slide .title[ # Semi-supervised Deep Learning ] .subtitle[ ## From autoencoders to Beta-VAEs ] .author[ ### <b>Marcin Kierczak</b> | 21-Mar-2023 ] .institute[ ### NBIS, SciLifeLab ] --- exclude: true count: false <link href="https://fonts.googleapis.com/css?family=Roboto|Source+Sans+Pro:300,400,600|Ubuntu+Mono&subset=latin-ext" rel="stylesheet"> <link rel="stylesheet" href="https://use.fontawesome.com/releases/v5.3.1/css/all.css" integrity="sha384-mzrmE5qonljUremFsqc01SB46JvROS7bZs3IO2EmfFsd15uHvIt+Y8vEf7N7fWAU" crossorigin="anonymous"> <!-- ------------ Only edit title, subtitle & author above this ------------ --> --- name: neural_zoo # Neural 🧠 Zoo 🐫 🐿 🦓 🐬 .pull-left-50[ .size-70[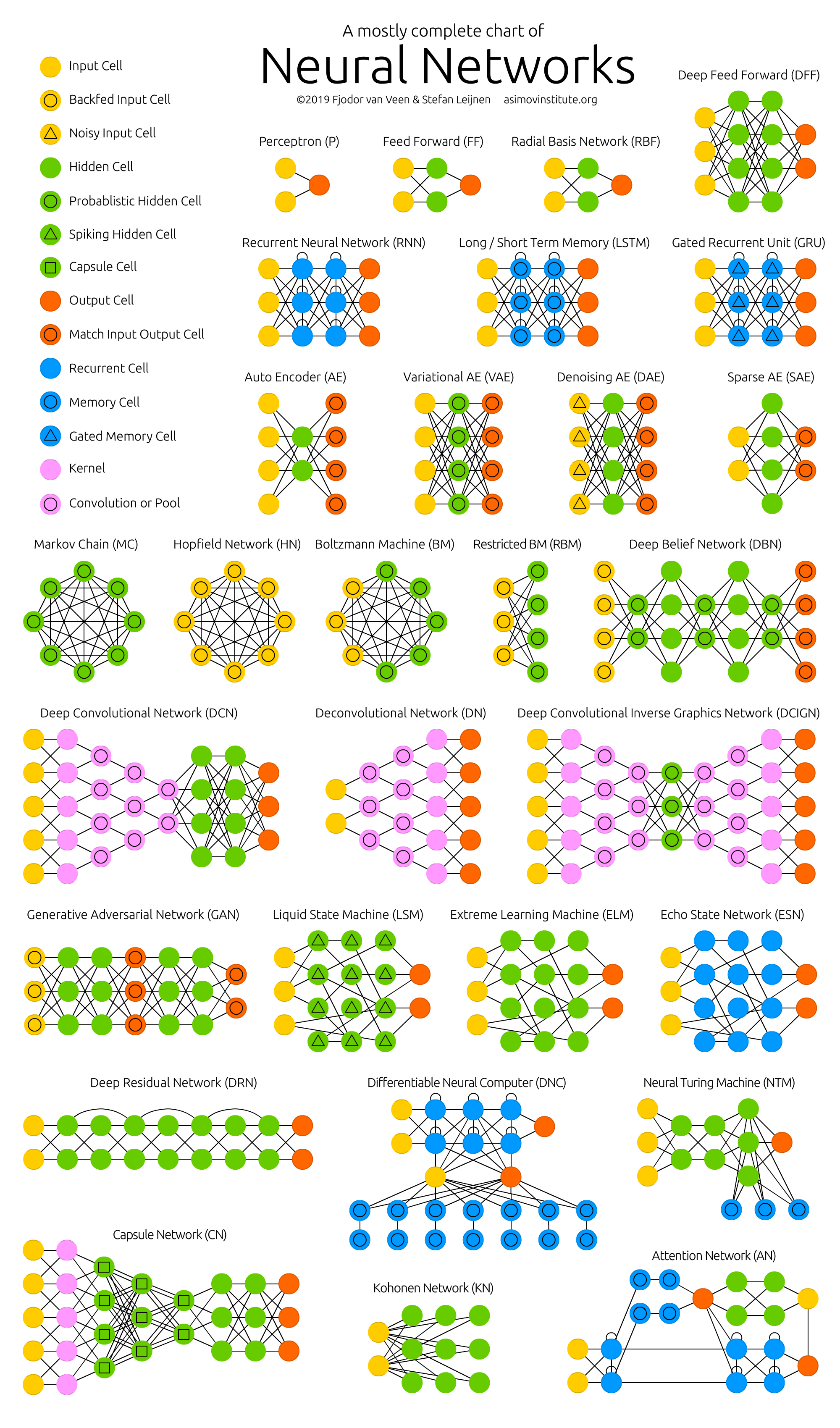] .small[source: [Asimov Institute](https://www.asimovinstitute.org/neural-network-zoo/)] ] .pull-right-50[ An autoencoder is "just" a specific ANN architecture: .size-35[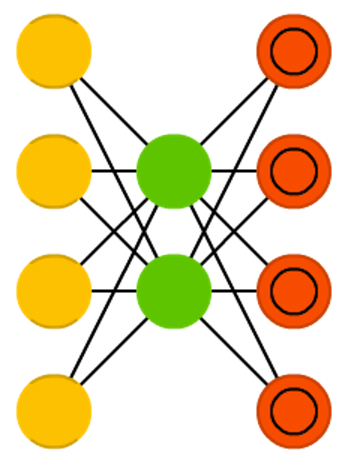] .small[source: [Asimov Institute](https://www.asimovinstitute.org/neural-network-zoo/)] ] --- name: autoencoders2 # Autoencoders So, what is really happening here❓ -- * output data (🎯) are THE SAME as the input data (not always, but a good assumption for now), -- * it is thus called **semi-supervised learning** (sometimes **self-supervised**), -- * the **loss function** favors weight sets that are able to reconstruct the data from itself ♻️, -- * why do we want a network that can reconstruct input data? A waste 🗑 of resources? -- * not really! What we want is the 🍾. .pull-left-50[ .size-70[] .small[source: [Wikimedia Commons](assets/autoencoder_schema.png)] ] --- name: applications_compression # Applications: compression .size-90[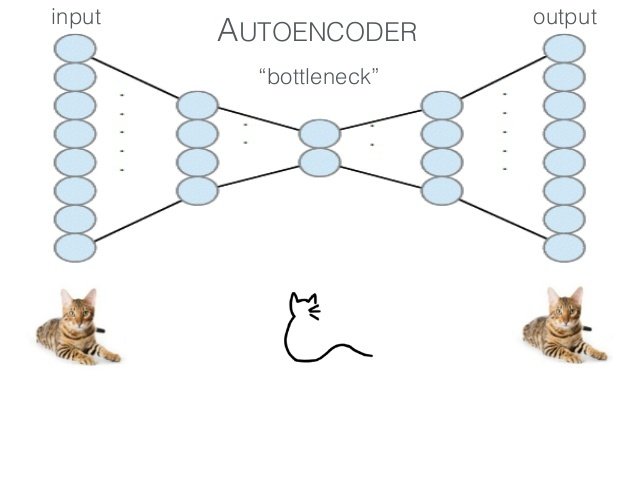] .small[source: [sefix.com](https://sefiks.com/2018/03/21/autoencoder-neural-networks-for-unsupervised-learning/)] --- name: denoising_autoenc # Applications: denoising 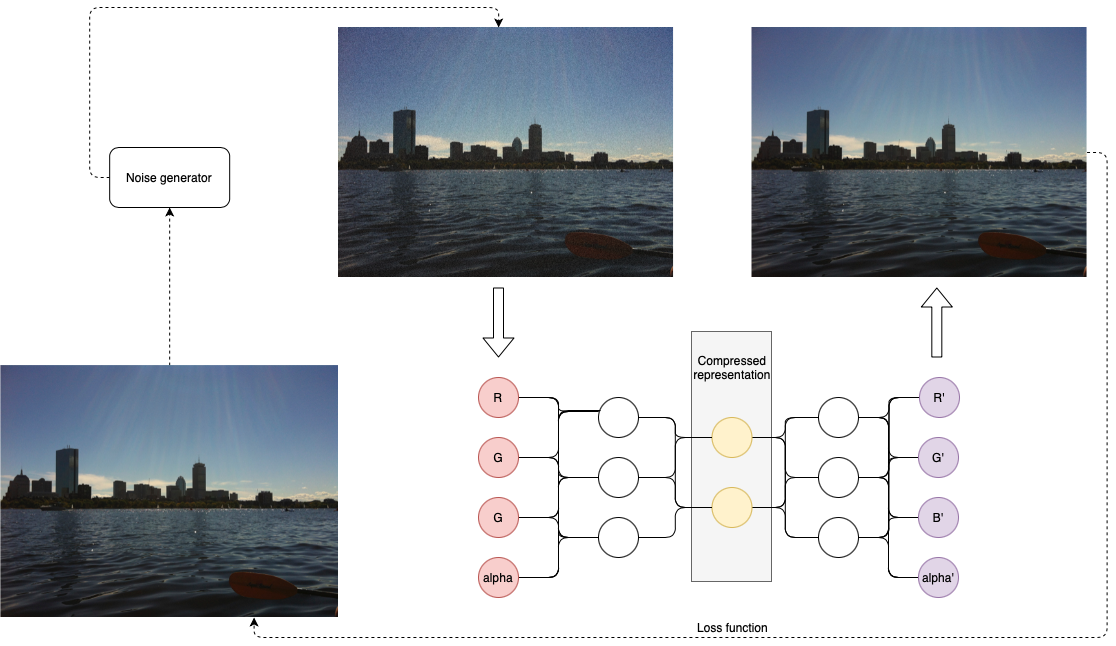 --- name: denoising_autoenc # Applications: neural inprinting  --- name: semantic_vae # Applications: semantic segmentation .pull-left-50[  ] .pull-right-50[  ] --- name: ae_discontinuity # Bad, bad, ugly latent space 💩 .size-60[] .small[source: [Irhum Shafkat](https://towardsdatascience.com/intuitively-understanding-variational-autoencoders-1bfe67eb5daf)] --- name: ae_vs_vae # AE vs. VAE latent space  --- name: vae1 # Variational AutoEncoder (VAE) .pull-left-50[ .small[ source: [Irhum Shafkat](https://towardsdatascience.com/intuitively-understanding-variational-autoencoders-1bfe67eb5daf) ] ] --- name: vae2 # Variational AutoEncoder (VAE) .pull-left-50[] .pull-right-50[] .small[source: [Irhum Shafkat](https://towardsdatascience.com/intuitively-understanding-variational-autoencoders-1bfe67eb5daf)] --- name: desired_latent_space # Towards latent space we want .size-60[] --- name: reparametrization_trick # The reparametrization trick 🎩🐰  .small[source: [antkillerfarm](http://antkillerfarm.github.io/images/img2/VAE_13.png)] --- name: vae2 # Loss function `\(\mathcal{L}(\Theta, \phi; \bar{x}, \bar{z}) = \mathbb{E}_{q_{\phi}(\bar{z}|\bar{x})}[\log{p_{\phi}}(\bar{x}|\bar{z})]\)` .size-80[] -- We need some 👮 👮! --- name: kl_div # Kullback-Leibler divergence Officers Solomon Kullback and Richard Leibler 🚔! --  -- > NOTE! `\(D_KL(P|Q) \neq D_KL(Q|P)\)` and that is why it is a divergence, not a distance. --- name: KL_in_R # Kullback-Leibler in R `\(D_{KL} = P(x) \times \log{\left(\frac{P(x)}{Q(x)}\right)}\)` ```r KL_div <- function(mu1 = 0, sigma1 = 1, mu2, sigma2) { g <- function(x) { P <- dnorm(x, mean=mu1, sd=sigma1, log=F) logP <- dnorm(x, mean=mu1, sd=sigma1, log=T) logQ <- dnorm(x, mean=mu2, sd=sigma2, log=T) val <- P * (logP - logQ) return(val) } result <- integrate(g, -Inf, Inf) return(result$value) } KL_div(mu1 = 0, sigma1 = 1, mu2 = 0, sigma2 = 1) KL_div(mu1 = 0, sigma1 = 1, mu2 = 0, sigma2 = 2) KL_div(mu1 = 0, sigma1 = 1, mu2 = 1, sigma2 = 1) ``` ``` ## [1] 0 ## [1] 0.3181472 ## [1] 0.5 ``` --- name: KL_only_loss # What if we only use `\(\mathcal{D}_{KL}\)`? `\(\mathcal{L}(\Theta, \phi; \bar{x}, \bar{z}) = \mathcal{D}_{KL}(q_{\phi}(\bar{z}|\bar{x})||p(\bar{z}))\)` The ⛪ 👨🎓 Bayesian connection... -- .size-50[] --- name: vae_full_loss_fn # Complete loss function `\(\mathcal{L}(\Theta, \phi; \bar{x}, \bar{z}) = \mathbb{E}_{q_{\phi}(\bar{z}|\bar{x})}[\log{p_{\phi}}(\bar{x}|\bar{z})] - \mathcal{D}_{KL}(q_{\phi}(\bar{z}|\bar{x})||p(\bar{z}))\)` .size-60[] --- name: beta-vae # `\(\beta\)`-VAEs We can make neurons in the latent layer (bottleneck) **disentangled**, i.e., force that they learn different generative parameters: `\(\mathcal{L}(\Theta, \phi; \bar{x}, \bar{z}) = \mathbb{E}_{q_{\phi}(\bar{z}|\bar{x})}[\log{p_{\phi}}(\bar{x}|\bar{z})] - \mathbf{\beta} \times \mathcal{D}_{KL}(q_{\phi}(\bar{z}|\bar{x})||p(\bar{z}))\)`  Image after: [β-VAE: Learning Basic Visual Concepts with a Constrained Variational Framework, Higgins et al., ICLR, 2017](https://openreview.net/pdf?id=Sy2fzU9gl) Further reading: [Understanding disentangling in β-VAE, Burgess et al., 2018](https://arxiv.org/abs/1804.03599) --- name: beta_vae_ex2 # `\(\beta\)`-VAEs .size-70[] .small[Source: [β-VAE: Learning Basic Visual Concepts with a Constrained Variational Framework, Higgins et al., ICLR, 2017](https://openreview.net/pdf?id=Sy2fzU9gl)] --- name: ae_vae_summary1 # Summary .size-70[] .small[source: [iart.ai](http://iart.ai)] --- name: ae_vae_summary2 # Summary * Autoencoders (AE) -- specific ANN architecture. * Autoencoders: + non-generative + `\(\mathcal{L}\)` minimizes reconstruction loss + latent space is discontinuous and has "gaps" + good enough for great many applications * Variational Autoencoders: + generative + `\(\mathcal{L}\)` minimizes reconstruction loss and **latent loss** + latent space has no gaps and is continuous + contain stochastic node -- need a reparametrization trick + still, the distributions are entangled and have no obvious meaning * `\(\beta\)`-VAEs (disentangled VAEs): + same as VAEs but the distributions have interpretation. --- name: topic1 ## If you need, `Keras` and `TensorFlow` in R * you need TensorFlow first, -- * than, you also need the `tensorflow` [package](https://tensorflow.rstudio.com/installation/)... -- * good 😀 news is you do not need to install manually... -- * install `keras` package instead. -- ```r install.packages('keras') keras::install_keras() ``` -- * if your graphics card supports CUDA and you want to use the power of GPU. ```r install.packages('keras') keras::install_keras(tensorflow = 'gpu') ``` -- * at least two packages provide R interfaces to `Keras`: `keras` by RStudio and `kerasR`. The latter does not expose all Keras functionalities to R though. --- name: topic2 ## R ❤️ Keras & TensorFlow There are excellent resources for learning: * about TensorFlow [package](https://tensorflow.rstudio.com/guide/tensorflow/eager_execution/) from RStudio, -- * excellent book by Chollet & Allaire .size-20[] -- * RStudio Deep Learning with Keras [cheatsheet](https://github.com/rstudio/cheatsheets/raw/master/keras.pdf) .size-20[] -- * from [keras.rstudio.com](https://keras.rstudio.com) <!-- --------------------- Do not edit this and below --------------------- --> --- name: end_slide class: end-slide, middle count: false # Let's build some autoencoders! .end-text[ <p>R version 4.2.2 (2022-10-31)<br><p>Platform: x86_64-apple-darwin17.0 (64-bit)</p><p>OS: macOS Big Sur ... 10.16</p><br> <hr> <span class="small">Built on : <i class='fa fa-calendar' aria-hidden='true'></i> 21-Mar-2023 at <i class='fa fa-clock-o' aria-hidden='true'></i> 21:57:13</span> <b>2023</b> • [SciLifeLab](https://www.scilifelab.se/) • [NBIS](https://nbis.se/) ]